crAssphage
| crAssphage | |
|---|---|
| |
| crAssphage | |
| Virus classification | |
| Group: | Group I (dsDNA)
|
| Order: | |
| Family: | |
| Subfamily: | |
| Genus: | Approximately 10 genera
|
| Type species | |
| |
crAssphage (cross-assembly phage) is a bacteriophage (virus that infects bacteria), discovered in 2014 by computational analysis of human faecal metagenomes[3] and later isolated for the first time in 2018.[4] It infects the human gut symbiont Bacteroides.[4][5]
crAssphage
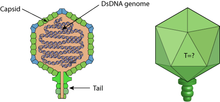
CrAssphage has a circular DNA genome 102 kbp in size and contains 80 predicted open reading frames, and the sequence is commonly found in human faecal samples. At the time of its discovery, the virus was predicted to infect bacteria of the phylum Bacteroidota that are common in the intestinal tract of many animals including humans.[3]
Based on analysis of metagenomics data, crAssphage sequences have been identified in about half of all sampled humans.[3] The virus was named after the crAss (cross-assembly)[6][7] software that was used to find the viral genome. CrAssphage is possibly the first organism to be named after a computer program for promotional purposes.[8]
CrAss-phage does not correlate strongly with variables relating to diet, lifestyle, or health, and does not appear to be strongly involved in human health or disease.[9][10] The virus may outperform indicator bacteria as a marker for human faecal contamination.[11][12][13]

The RNA polymerase of crAssphage is homologous to proteins catalyzing RNA interference in animals and humans, rather than to their own RNA polymerase.[14]
crAss-like phages
In silico comparative genomics and taxonomic analysis have found that crAss-like phages represent a highly abundant family of viruses. Based on a screen of related sequences in public nucleotide databases and phylogenetic analysis, it was concluded that crAssphage may be part of an extensive bacteriophage group (within Podoviridae, order Caudovirales) with diverse members that are found in a range of environments including human gut and faeces, termite gut,[1][15][16][17] terrestrial/groundwater environments, soda lake (hypersaline brine), marine sediment, and plant root environments.[1] CrAss-like phages are estimated to comprise about 10 genera.[18]
Since then, so-called gubaphages have been identified as the second most common phage group in the human intestinal flora. The characteristics of the gubaphages are reminiscent of those of "p-crAssphage".[19][20]
References
- ^ a b c d e Yutin N, Makarova KS, Gussow AB, Krupovic M, Segall A, Edwards RA, Koonin EV (January 2018). "Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut". Nature Microbiology. 3 (1): 38–46. doi:10.1038/s41564-017-0053-y. PMC 5736458. PMID 29133882.; and Eugene V. Koonin: Behind the paper: The most abundant human-associated virus no longer an orphan, November 13th, 2017
- ^ NCBI: crAss-like viruses/phages (clade)
- ^ a b c Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, Boling L, et al. (July 2014). "A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes". Nature Communications. 5: 4498. Bibcode:2014NatCo...5.4498D. doi:10.1038/ncomms5498. PMC 4111155. PMID 25058116.
- ^ a b Shkoporov AN, Khokhlova EV, Fitzgerald CB, Stockdale SR, Draper LA, Ross RP, Hill C (November 2018). "ΦCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis". Nature Communications. 9 (1): 4781. doi:10.1038/s41467-018-07225-7. PMC 6235969. PMID 30429469.
- ^ Guerin E, Shkoporov AN, Stockdale SR, Comas JC, Khokhlova EV, Clooney AG, et al. (April 2021). "Isolation and characterisation of ΦcrAss002, a crAss-like phage from the human gut that infects Bacteroides xylanisolvens". Microbiome. 9 (1): 89. doi:10.1186/s40168-021-01036-7. PMC 8042965. PMID 33845877.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Cross-Assembly of Metagenomes
- ^ Dutilh BE, Schmieder R, Nulton J, Felts B, Salamon P, Edwards RA, Mokili JL (December 2012). "Reference-independent comparative metagenomics using cross-assembly: crAss". Bioinformatics. 28 (24): 3225–3231. doi:10.1093/bioinformatics/bts613. PMC 3519457. PMID 23074261.
- ^ Chamary JV. "A Common Virus Is Eating Your Gut Bacteria". Forbes. Retrieved 2019-04-19.
- ^ Kirsch JM, Brzozowski RS, Faith D, Round JL, Secor PR, Duerkop BA (September 2021). "Bacteriophage-Bacteria Interactions in the Gut: From Invertebrates to Mammals". Annual Review of Virology. 8 (1): 95–113. doi:10.1146/annurev-virology-091919-101238. PMC 8484061. PMID 34255542.
{{cite journal}}: CS1 maint: PMC embargo expired (link) - ^ Liang YY, Zhang W, Tong YG, Chen SP (December 2016). "crAssphage is not associated with diarrhoea and has high genetic diversity". Epidemiology and Infection. 144 (16): 3549–3553. doi:10.1017/S095026881600176X. PMID 30489235.
- ^ Ahmed W, Lobos A, Senkbeil J, Peraud J, Gallard J, Harwood VJ (March 2018). "Evaluation of the novel crAssphage marker for sewage pollution tracking in storm drain outfalls in Tampa, Florida". Water Research. 131: 142–150. doi:10.1016/j.watres.2017.12.011. PMID 29281808.
- ^ García-Aljaro C, Ballesté E, Muniesa M, Jofre J (November 2017). "Determination of crAssphage in water samples and applicability for tracking human faecal pollution". Microbial Biotechnology. 10 (6): 1775–1780. doi:10.1111/1751-7915.12841. PMC 5658656. PMID 28925595.
- ^ Stachler E, Kelty C, Sivaganesan M, Li X, Bibby K, Shanks OC (August 2017). "Quantitative CrAssphage PCR Assays for Human Fecal Pollution Measurement". Environmental Science & Technology. 51 (16): 9146–9154. Bibcode:2017EnST...51.9146S. doi:10.1021/acs.est.7b02703. PMC 7350147. PMID 28700235.
- ^ Gut microbiome manipulation could result from virus discovery. On: EurekAlert! 18 November 2020. Source: Rutgers University
- ^ Tikhe CV, Husseneder C (2018). "Metavirome Sequencing of the Termite Gut Reveals the Presence of an Unexplored Bacteriophage Community". Frontiers in Microbiology. 8: 2548. doi:10.3389/fmicb.2017.02548. PMC 5759034. PMID 29354098.
- ^ Pramono AK, Kuwahara H, Itoh T, Toyoda A, Yamada A, Hongoh Y (June 2017). "Discovery and Complete Genome Sequence of a Bacteriophage from an Obligate Intracellular Symbiont of a Cellulolytic Protist in the Termite Gut". Microbes and Environments. 32 (2): 112–117. doi:10.1264/jsme2.ME16175. PMC 5478533. PMID 28321010.
- ^ Guerin E, Shkoporov A, Stockdale SR, Clooney AG, Ryan FJ, Sutton TD, et al. (November 2018). "Biology and Taxonomy of crAss-like Bacteriophages, the Most Abundant Virus in the Human Gut". Cell Host & Microbe. 24 (5): 653–664.e6. doi:10.1016/j.chom.2018.10.002. PMID 30449316. S2CID 53950837.
- ^ SIB: crAsslike phages, Expasy ViralZone
- ^ Luis Fernando Camarillo-Guerrero, Alexandre Almeida, Guillermo Rangel-Pineros, Robert D. Finn, Trevor D. Lawley: Massive expansion of human gut bacteriophage diversity. In: Cell Resource Volume 184, No. 4, pp 1098-1109.e9, 18 February 2021, doi:10.1016/j.cell.2021.01.029. PrePrint as of 3 Sep. 2020: bioRxiv, Europe PMC, doi:10.1101/2020.09.03.280214. See also:
- Biologists Find Almost 143,000 Bacteriophage Species in Human Gut, auf: sci-news vom 19. Februar 2021 (englisch)
- ^ Luis Fernando Camarillo Guerrero: Integrative Analysis of the Human Gut Phageome Using a Metagenomics Approach, Thesis, Gonville & Caius College, University of Cambridge, August 2020, doi:10.17863/CAM.63973
